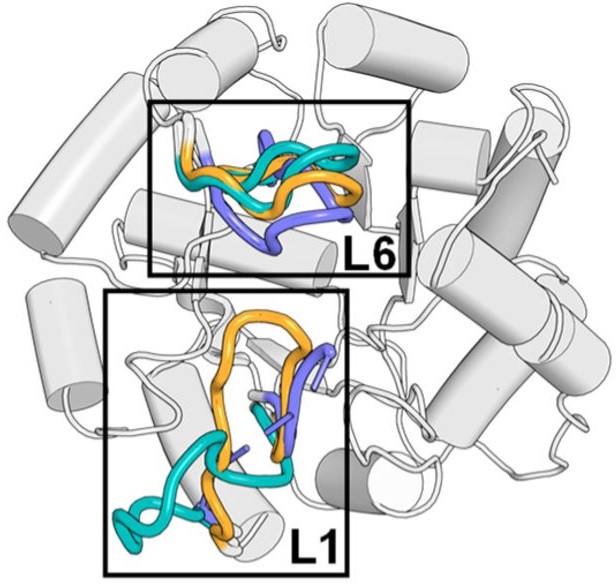
Distal mutations in a designed retro-aldolase alter loop dynamics to shift and accelerate the rate-limiting step Hunt SE, Klaus C, John AE, Zarifi N, Martinez A, Feixas F, Garcia-Borras M, Thompson MC, Chica RA. (2024) bioRxiv.”
- PMID:
- Biorxiv Preprint: 2025.01.26.634918
- Full Text
- Deposited Structures: 9MYA, 9MYB
- Chica Lab @ UOttawa
- Garcia-Borras Lab @ iQCC
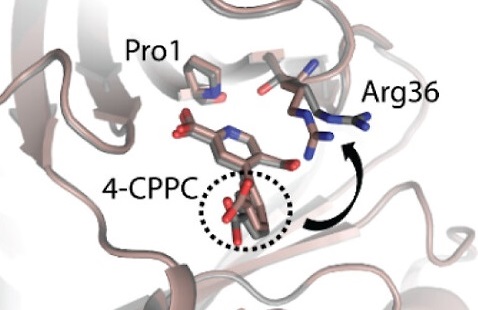
The C-terminal Region of D-DT Regulates Molecular Recognition for Protein–Ligand Complexes
Parkins A, Pilien AVR, Wolff AM, Argueta C, Vargas J, Sadeghi S, Franz AH, Thompson MC, Pantouris G. (2024) Journal of Medicinal Chemistry.”
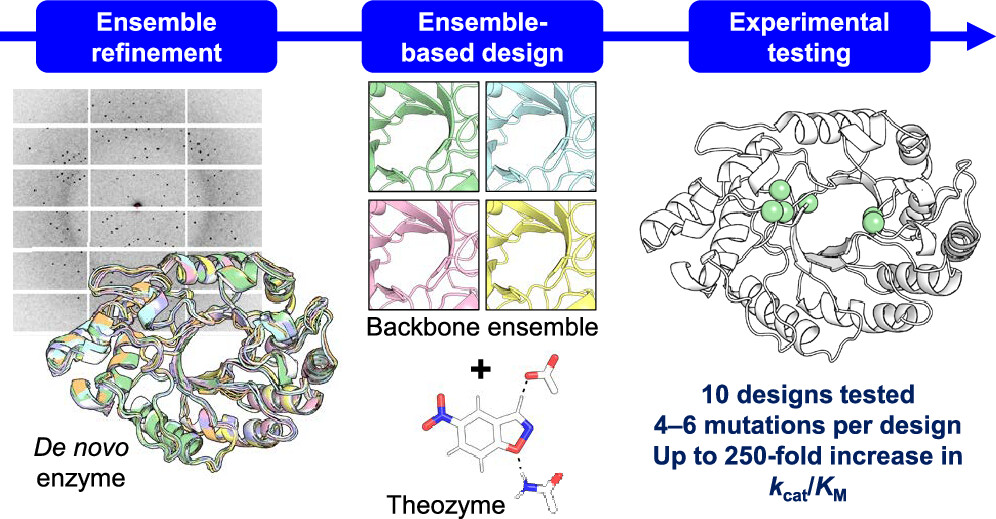
Design of Efficient Artificial Enzymes Using Crystallographically Enhanced Backbone Sampling
Rakotoharisoa RV, Seifinoferest B, Zarifi N, Miller JDM, Rodriguez JM, Thompson MC, Chica RA. (2024) JACS.”
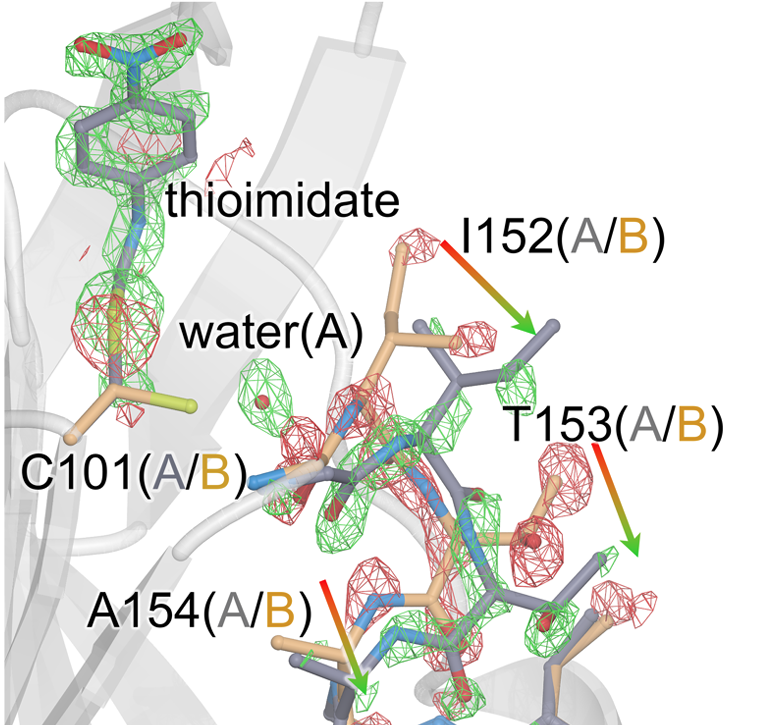
Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography
Smith N, Dasgupta M, Wych DC, Dolamore C, Sierra RG, Lisova S, Marchany-Rivera D, Cohen AE, Boutet S, Hunter MS, Kupitz C, Poitevin F, Moss FR III, Mittan-Moreau DW, Brewster AS, Sauter NK, Young ID, Wolff AM, Tiwari VK, Kumar N, Berkowitz DB, Hadt RG, Thompson MC, Follmer AH, Wall ME, Wilson MA. (2024) Science Advances.”
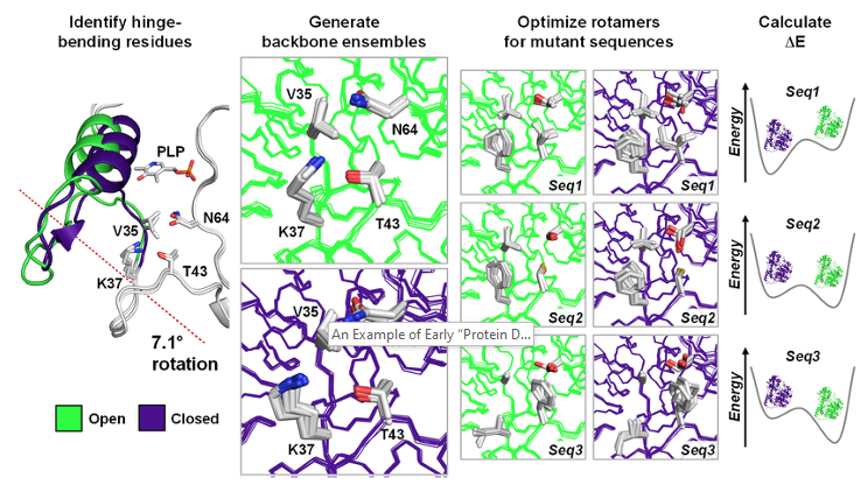
Computational remodeling of an enzyme conformational landscape for altered substrate selectivity
St-Jacques AD, Rodriguez JM, Eason MG, Foster SM, Khan ST, Damry AM, Goto NK, Thompson MC, Chica RA. (2023) Nature Communications.
- PMID: 37770431
- PMCID: PMC10539519
- Biorxiv Preprint: 2022.09.16.508321
- Full Text
- Deposited Structures: 8E9P, 8E9K, 8E9J, 8E9Q, 8E9L, 8E9M, 8E9N, 8E9O, 8E9R, 8E9S, 8E9C, 8E9D
- Chica Lab @ UOttawa
- Goto Lab @ UOttawa
- Damry Lab @ UOttawa
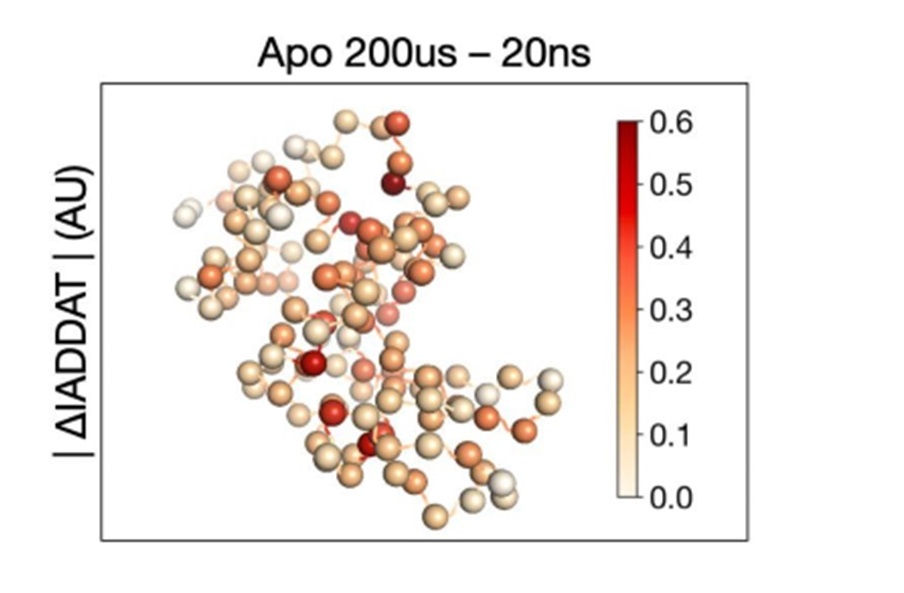
Mapping Protein Dynamics at High Spatial Resolution with Temperature-Jump X-ray Crystallography
Wolff AM, Nango E, Young ID, Brewster AS, Kubo M, Nomura T, Sugahara M, Owada S, Barad BA, Ito K, Bhowmick A, Carbajo S, Hino T, Holton JM, Im D, O’Riordan LJ, Tanaka T, Tanaka R, Sierra RG, Yumoto F, Tono K, Iwata S, Sauter NK, Fraser JS, Thompson MC. (2023) Nature Chemistry.
- PMID: 37723259
- PMCID: PMC10624634
- Biorxiv Preprint: 2022.06.10.495662
- Full Text
- Online Dataset: doi:10.11577/1873469
- Deposited Structures: 8CVU, 8CVV, 8CVW, 8CW0, 8CW1, 8CW3, 8CW5, 8CW6, 8CW7, 8CW8, 8CWB, 8CWC, 8CWD, 8CWE, 8CWF, 8CWG, 8CWH
- In the Pipeline by Derek Lowe
- GitHub Repository
- Nango lab @ Tohoku University
- Fraser lab @ UC San Francisco
- SACLA XFEL
- BioSciences @ LBNL
- Preprint Tweetstorm
Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase
Diaz RE, Ecker AK, Correy GJ, Asthana P, Young ID, Faust B, Thompson MC, Seiple IB, Van Dyken SJ, Locksley RM, Fraser JS. (2023) eLife.
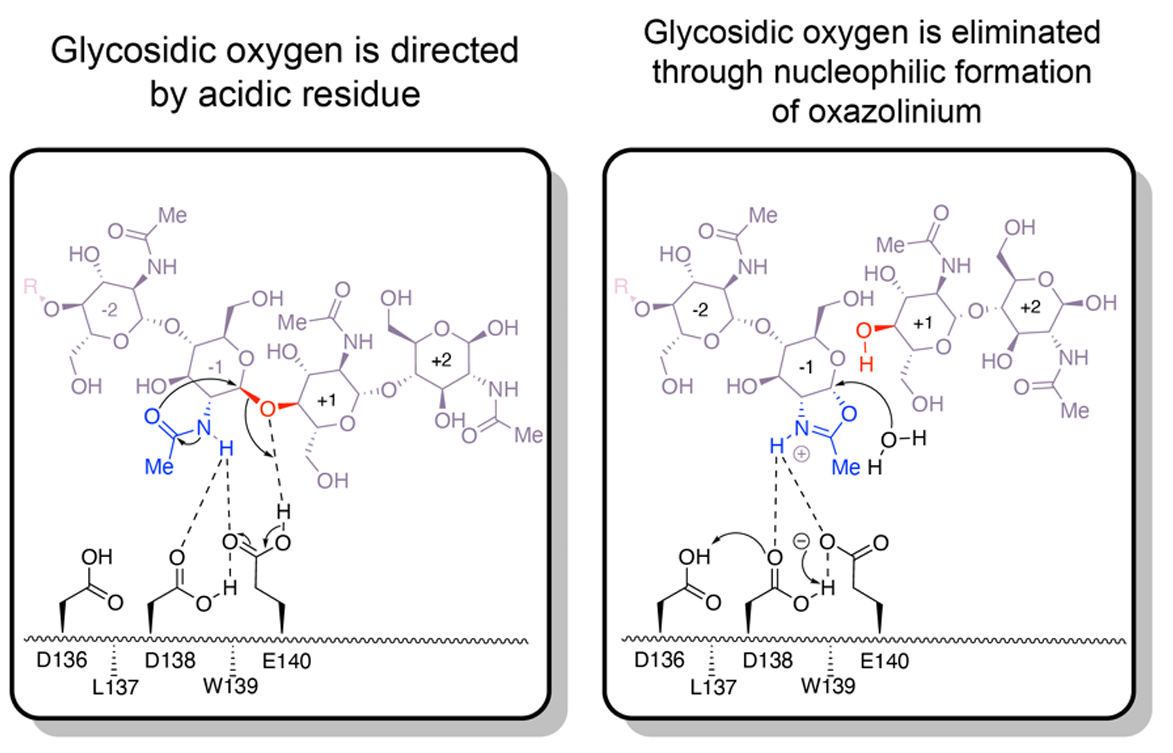
- PMID:
- Biorxiv Preprint: 2023.06.03.542675
- Full Text
- Deposited Structures: 8FG5, 8FG7, 8GCA, 8FRC, 8FR9, 8FRB, 8FRD, 8FRG, 8FRA
- Fraser Lab @ UC San Francisco
- Seiple Lab @ UC San Francisco
- Van Dyken Lab @ Washington U in St. Louis
- Locksley Lab @ UC San Francisco
Combining temperature perturbations with X-ray crystallography to study dynamic macromolecules: A thorough discussion of experimental methods
Thompson MC (2023) Methods in Enzymology.
The mechanisms of catalysis and ligand binding for the SARS-CoV-2 NSP3 macrodomain from neutron and X-ray diffraction at room temperature
Correy GJ, Kneller DW, Phillips G, Pant S, Russi S, Cohen AE, Meigs G, Holton JM, Gahbauer S, Thompson MC, Ashworth A, Coates L, Kovalevsky A, Meilleur F, Fraser JS. (2022) Science Advances.
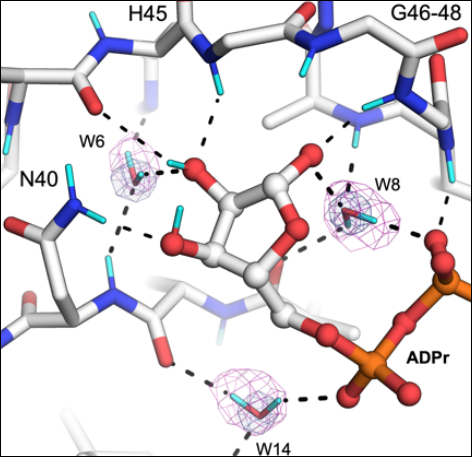
- PMID: 35622909
- PMCID: PMC9140965
- Biorxiv Preprint: 2022.02.07.479477
- Full Text
- Fraser Lab @ UC San Francisco
- Ashworth Lab @ UC San Francisco
- Meilleur Lab @ NC State University
- Oak Ridge National Lab
- Celebratory Tweetstorm by James Fraser
Accurately positioning functional residues with robotics-inspired computational protein design
Krivacic C, Kundert K, Pan X, Pache RA, Liu L, Conchuir SO, Jeliazkov JR, Gray JJ, Thompson MC, Fraser JS, Kortemme T. (2022) PNAS.
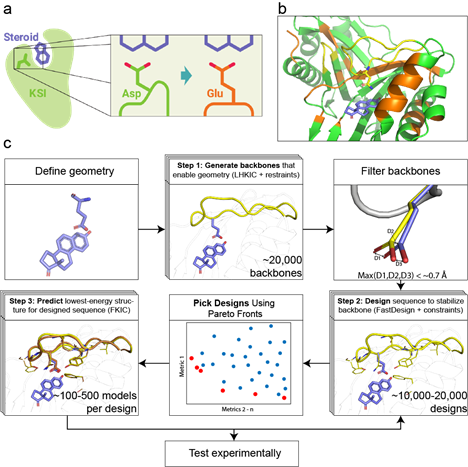
- PMID: 35254891
- PMCID: PMC8931229
- Biorxiv Preprint: 2021.07.02.450934
- Full Text
- Kortemme lab @ UC San Francisco
- Fraser lab @ UC San Francisco
- Gray lab @ Johns Hopkins U
Generation of bright monomeric red fluorescent proteins via computational design of enhanced chromophore packing
Legault S, Fraser-Halberg DP, McAnelly RL, Eason MG, Thompson MC, Chica RA. (2022) Chemical Science.
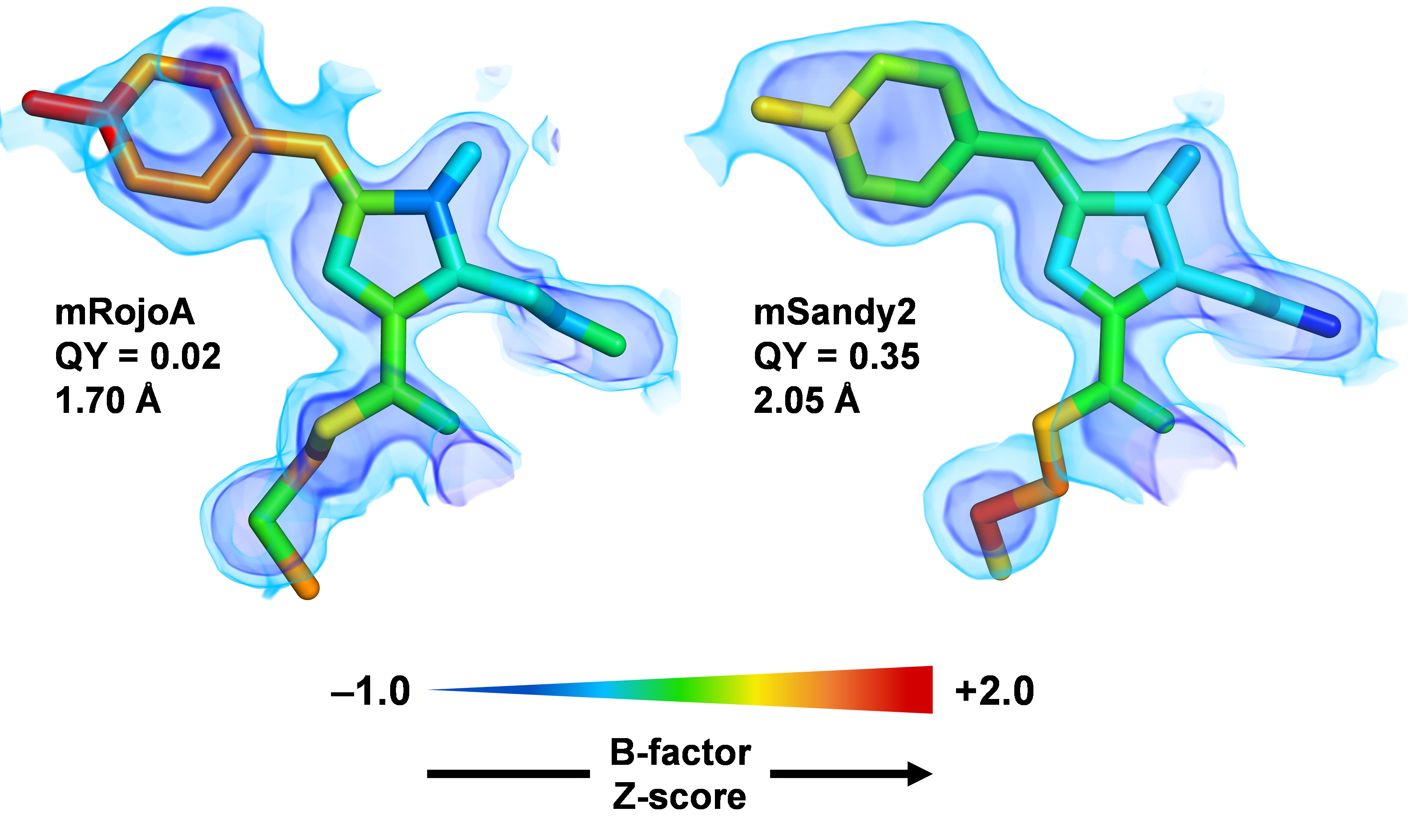
- PMID: 35222925
- PMCID: PMC8809391
- Full Text
- Chica Lab @ UOttawa
Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking
Schuller M, Correy GJ, Gahbauer S, Fearon D, Wu T, Diaz RE, Young ID, Martins LC, Smith DH, Schulze-Gahmen U, Owens TW, Deshpande I, Merz GE, Thwin AC, Biel JT, Peters JK, Moritz M, Herrera N, Kratochvil HT, QCRG Structural Biology Consortium, Aimon A, Bennett JM, Brandao Neto J, Cohen AE, Dias A, Douangamath A, Dunnett L, Fedorov O, Ferla MP, Fuchs M, Gorrie-Stone TJ, Holton JM, Johnson MG, Krojer T, Meigs G, Powell AJ, Rack JGM, Rangel VL, Russi S, Skyner RE, Smith CA, Soares AS, Wierman JL, Zhu K, Jura N, Ashworth A, Irwin J, Thompson MC, Gestwicki JE, von Delft F, Shoichet BK, Fraser JS, Ahel I. (2021) Science Advances.
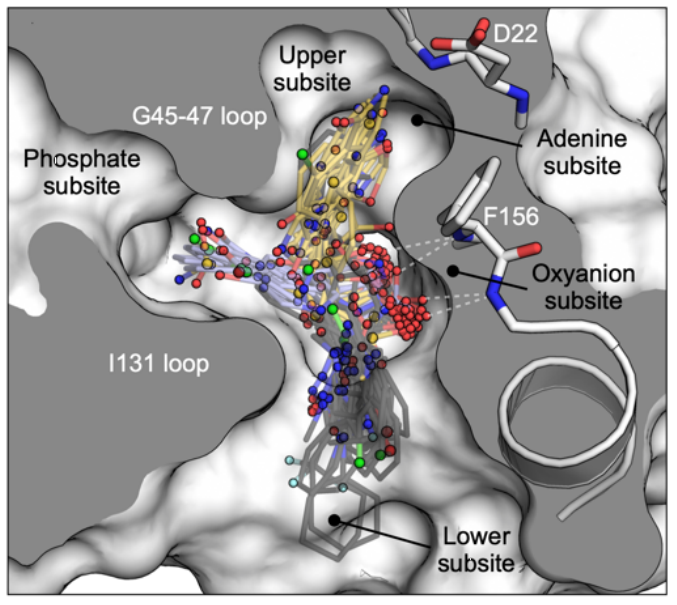
- PMID: 33853786
- PMCID: PMC8046379
- Biorxiv Preprint: 2020.11.24.393405
- Full Text
- View structures of fragment hits with Fragalysis
- Ahel Lab @ Oxford
- Fraser Lab @ UCSF
- Shoichet Lab @ UCSF
- XChem @ Diamond Light Source
- Gestwicki Lab @ UCSF
- Ashworth Lab @ UCSF
- QCRG Structural Biology Consortium
- ALS Beamline 8.3.1
- Biosciences Division @ LBNL
- SSRL Structural Molecular Biology Beamlines @ SLAC
- FMX Beamline @ NSLS-II
An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike
Schoof M, Faust B, Saunders RA, Sangwan S, Rezelj VV, Hoppe N, Boone M, Billesboelle C, Puchades C, Azumaya CM, Kratochvil HT, Zimanyi M, Deshpande I, Liang J, Dickinson S, Nguyen HC, Chio CM, Merz GE, Thompson MC, Diwanji D, Schaefer K, Anand AA, Dobzinski N, Zha BS, Simoneau CR, Leon K, White KM, Chi US, Gupta M, Jin M, Li F, Liu Y, Zhang K, Bulkley D, Sun M, Smith AM, Rizo AN, Moss F, Brilot AF, Pourmal S, Trenker R, Pospiech T, Gupta S, Barsi-Rhyne B, Belyy V, Barile-Hill AW, Nock S, Liu Y, Krogan NJ, Ralston CY, Swaney DL, Garcia-Sastre A, Ott M, Vignuzzi M, Quantitative Biosciences Institute (QBI) Coronavirus Research Group Structural Biology Consortium, Walter P, Manglik A. (2020) Science.
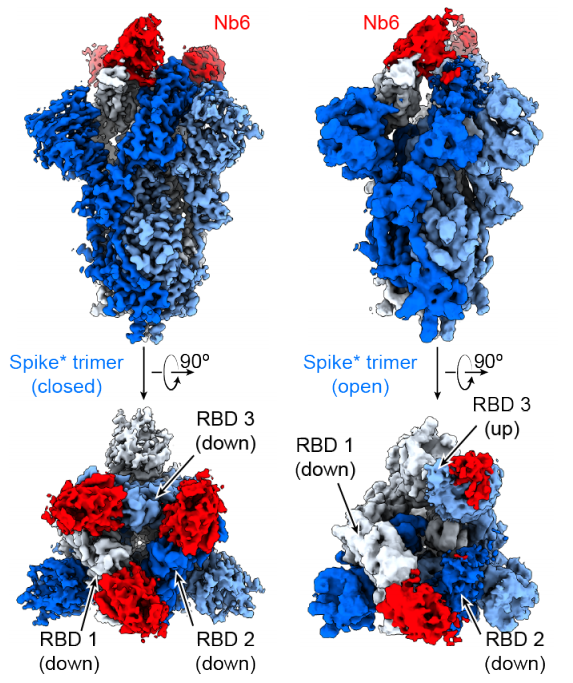
- PMID: 33154106
- PMCID: PMC7857409
- Biorxiv Preprint: 2020.08.08.238469
- Full Text
- QCRG Structural Biology Consortium
- Manglick Lab @ UCSF
- Walter Lab @ UCSF
- Vignuzzi Lab @ Institut Pasteur
- Ott Lab @ UCSF
- Garcia-Sastre Lab @ Mount Sinai
- Krogan Lab @ UCSF
- Biosciences Division @ LBNL
- UCSF News Release
- Coverage by Vice News
Ensemble-based enzyme design can recapitulate the effects of directed evolution in silico
Broom A, Rakotoharisoa RV, Thompson MC, Zarifi N, Nguyen E, Mukhametzhanov N, Liu L, Fraser JS, Chica RA. (2020) Nature Communications.
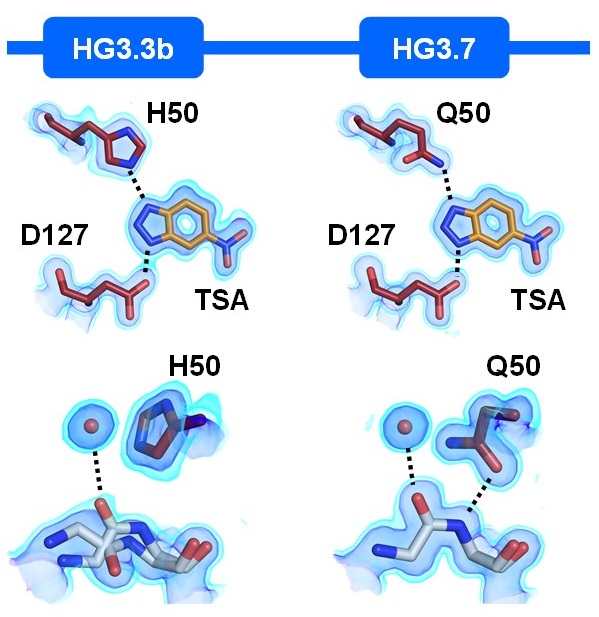
- PMID: 32968058
- PMCID: PMC7511930
- Biorxiv Preprint: 2020.03.19.999235
- Full Text
- Chica Lab @ UOttawa
- Fraser lab @ UC San Francisco
Expanding the space of protein geometries by computational design of de novo fold families
Pan X, Thompson MC, Zhang Y, Liu L, Fraser JS, Kelly MJS, Kortemme T. (2020) Science.
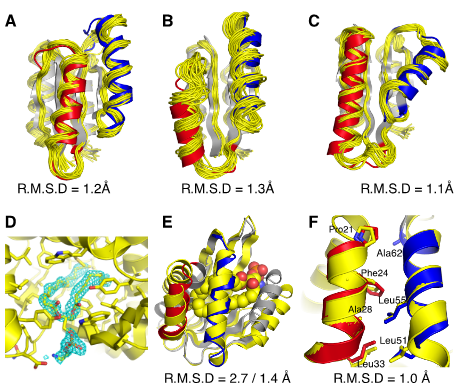
- PMID: 32855341
- PMCID: PMC7787817
- Biorxiv Preprint: 2020.04.14.041772
- Full Text
- Kortemme lab @ UC San Francisco
- Fraser lab @ UC San Francisco
Advances in methods for atomic resolution macromolecular structure determination
Thompson MC, Yeates TO, Rodriguez JA. (2020) F1000Res.
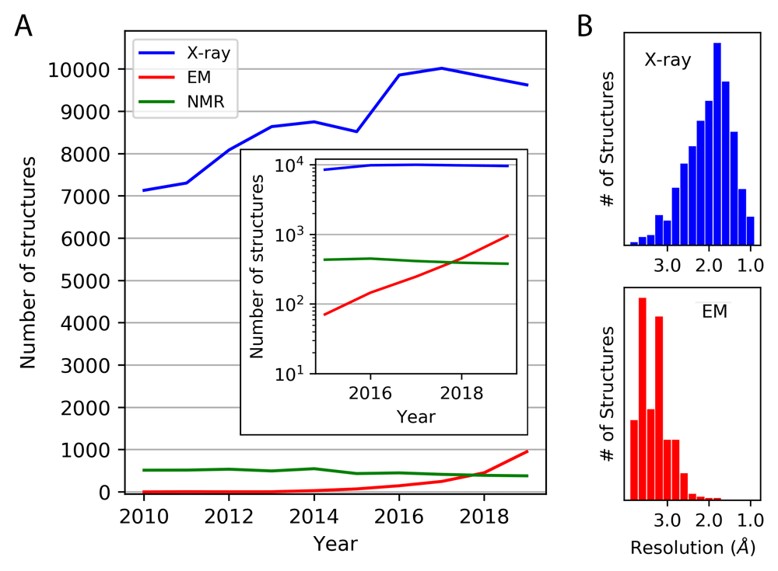
- PMID: 32676184
- PMCID: PMC7333361
- Full Text
- Yeates Lab @ UCLA
- Rodriguez Lab @ UCLA
Comparing serial X-ray crystallography and microcrystal electron diffraction (MicroED) as methods for routine structure determination from small macromolecular crystals
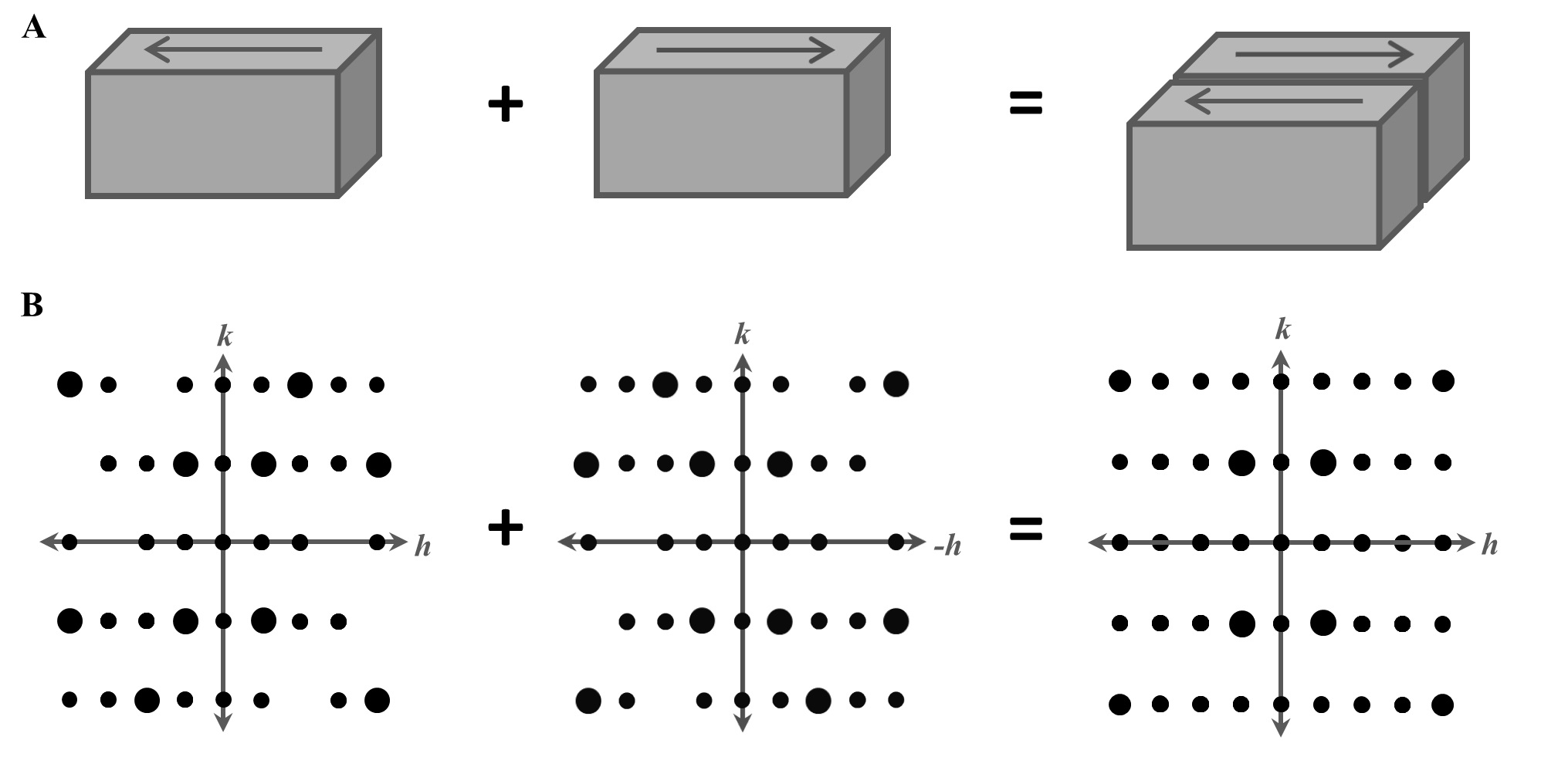
Wolff AM, Young ID, Sierra RG, Brewster AS, Martynowycz MW, Nango E, Sugahara M, Nakane T, Ito K, Aquila A, Bhowmick A, Biel JT, Carbajo S, Cohen AE, Cortez S, Gonzalez A, Hino T, Im D, Koralek JD, Kubo M, Lazarou TS, Nomura T, Owada S, Samelson A, Tanaka R, Tanaka T, Thompson EM, van den Bedem H, Woldeyes RA, Yumoto F, Zhao W, Tono K, Boutet S, Iwata S, Gonen T, Sauter NK, Fraser JS, Thompson MC. (2020) IUCrJ.
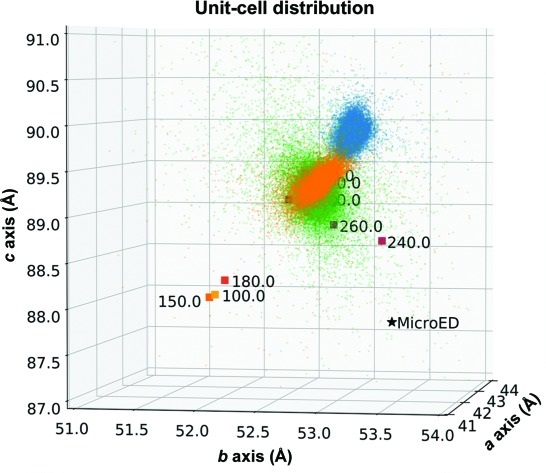
- PMID: 32148858
- PMCID: PMC7055375
- Biorxiv Preprint: 767061
- Full Text
- Online Dataset:
- Deposited Structures: 6U5C, 6U5D, 6U5E
- Fraser lab @ UC San Francisco
- Gonen lab @ UCLA
- LCLS MFX Beamline
- SACLA XFEL
- Celebratory Tweetstorm by corresponding author Michael Thompson
Mix-and-inject XFEL crystallography reveals gated conformational dynamics during enzyme catalysis
Dasgupta M, Budday D, Oliveira SHP, Madzelan P, Marchany-Rivera D, Seravalli J, Hayes B, Sierra RG, Boutet S, Hunter MS, Alonso-Mori R, Batyuk A, Wierman J, Lyubimov A, Brewster AS, Sauter NK, Applegate GA, Tiwari VK, Berkowitz DB, Thompson MC, Cohen AE, Fraser JS, Wall ME, van den Bedem H, Wilson MA. (2019) PNAS.
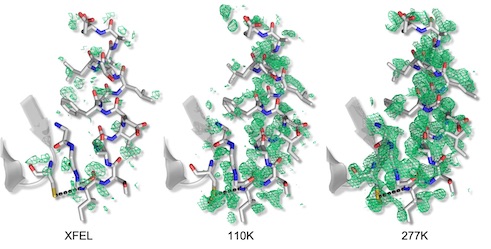
- PMID: 31801874
- PMCID: PMC6926069
- Biorxiv Preprint: 524751
- Full Text
- SLAC - Researchers reveal how enzyme motions catalyze reactions
- Enzymes and X-rays - Study reveals hidden acrobatics of cellular catalysts
- Wilson lab
- Henry van den Bedem
- Fraser lab @ UC San Francisco
- Paper Submission Celebration Tweets
Computational design of a modular protein sense/response system
Glasgow AA, Huang Y, Mandell DJ, Thompson M, Ritterson R, Loshbaugh AL, Pellegrino J, Krivacic C, Pache RA, Barlow KA, Ollikainen N, Jeon D, Kelly MJS, Fraser JS, Kortemme T. (2019) Science.
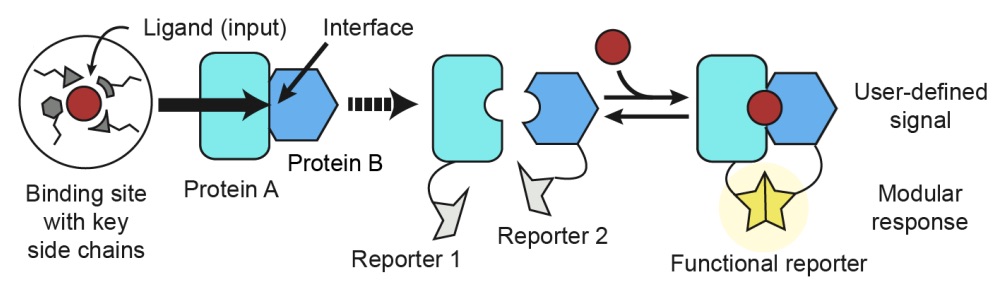
- PMID: 31754004
- Biorxiv Preprint: 648485
- Full Text
- Deposited Structure: 6OB5
- Perspective - Designer sense-response systems
- SBGrid - Computational Design of Protein Sensors Coupled with Functional Outputs
- Kortemme lab
- Fraser lab @ UC San Francisco
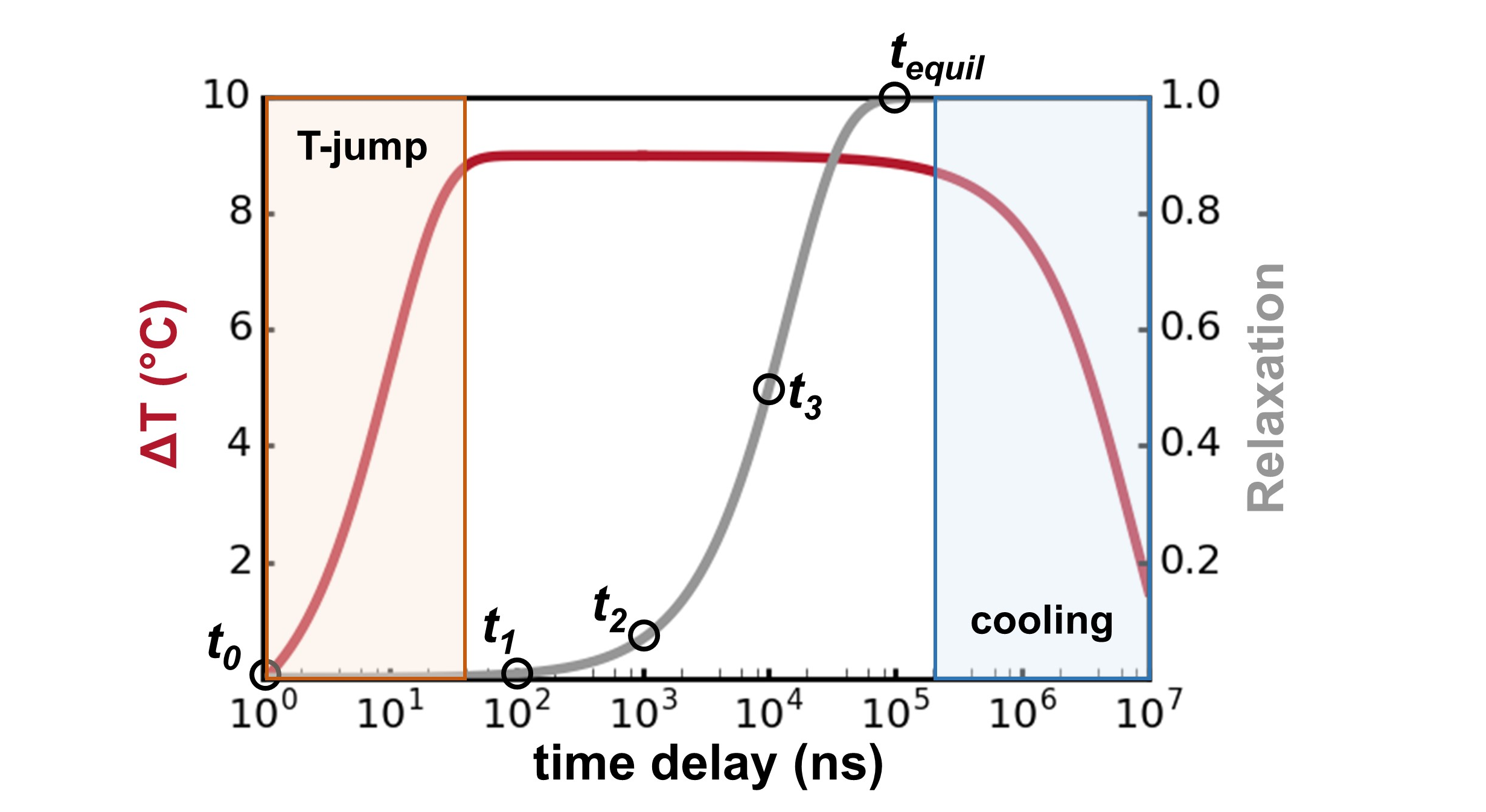
Temperature-Jump Solution X-ray Scattering Reveals Distinct Motions in a Dynamic Enzyme
Thompson MC, Barad BA, Wolff AM, Cho HS, Schotte F, Schwarz DMC, Anfinrud P, Fraser JS. (2019) Nature Chemistry.
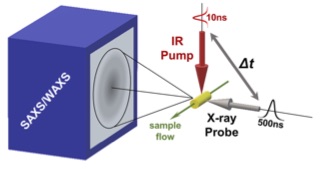
- PMID: 31527847
- PMCID: PMC6815256
- Biorxiv Preprint: 476432
- Full Text
- Solution Scattering Code on Github
- Solution Scattering Data on NIH Figshare
- Fraser lab @ UC San Francisco
- Philip Anfinrud
- BioCARS
- Paper Submission Celebration Tweetstorm
Microfocus diffraction from different regions of a protein crystal: structural variations and unit-cell polymorphism
Thompson MC, Cascio D, Yeates TO. (2018) Acta Cryst D.
- PMID: 29717712
- PMCID: PMC5930349
- Full Text
- Deposited Structures: 4TLH, 6ARC, 6ARD
- Yeates lab
Allosteric Inhibitors, Crystallography and Comparative Analysis Reveal Network of Coordinated Movement Across Human Herpesvirus Proteases
Acker TM, Gable JE, Bohn MF, Jaishankar P, Thompson MC, Fraser JS, Renslo AR, Craik CS. (2017) JACS.
Identifying and Overcoming Crystal Pathologies: Disorder and Twinning
Thompson MC. (2017) Protein Crystallography: Methods and Protocols.
Flexibility and design: conformational heterogeneity along the evolutionary trajectory of a redesigned ubiquitin
Biel JT, Thompson MC, Cunningham CN, Corn JE, Fraser JS. (2017) Structure.
- PMID: 28416112
- PMCID: PMC5415430
- Biorxiv Preprint: 081646
- Full Text
- Online Dataset:
- Deposited Structures: 5TOF, 5TOG
- Fraser lab @ UC San Francisco
- Paper Submission Celebration Photo
- Genentech
- Preview: Room Temperature X-Ray Crystallography Reveals Conformational Heterogeneity of Engineered Proteins
Structure of a novel 13 nm dodecahedral nanocage assembled from a redesigned bacterial microcompartment shell protein
Jorda J, Leibly DJ, Thompson MC, Yeates TO. (2016) Chemical Communications.
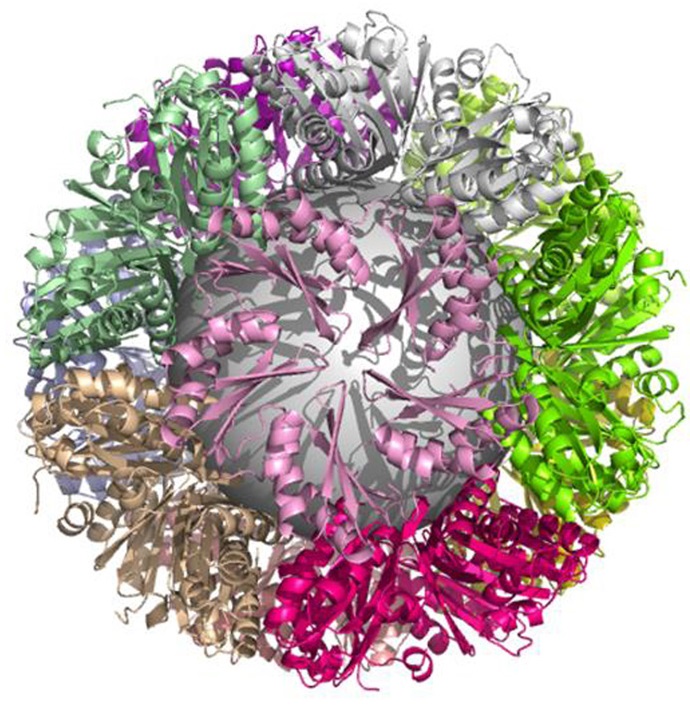
- PMID: 26988700
- PMCID: PMC5081076
- Full Text
- Deposited Structure: 5HPN
- Yeates lab
An allosteric model for control of pore opening by substrate binding in the EutL microcompartment shell protein
Thompson MC, Cascio D, Leibly DJ, Yeates TO. (2015) Protein Science.
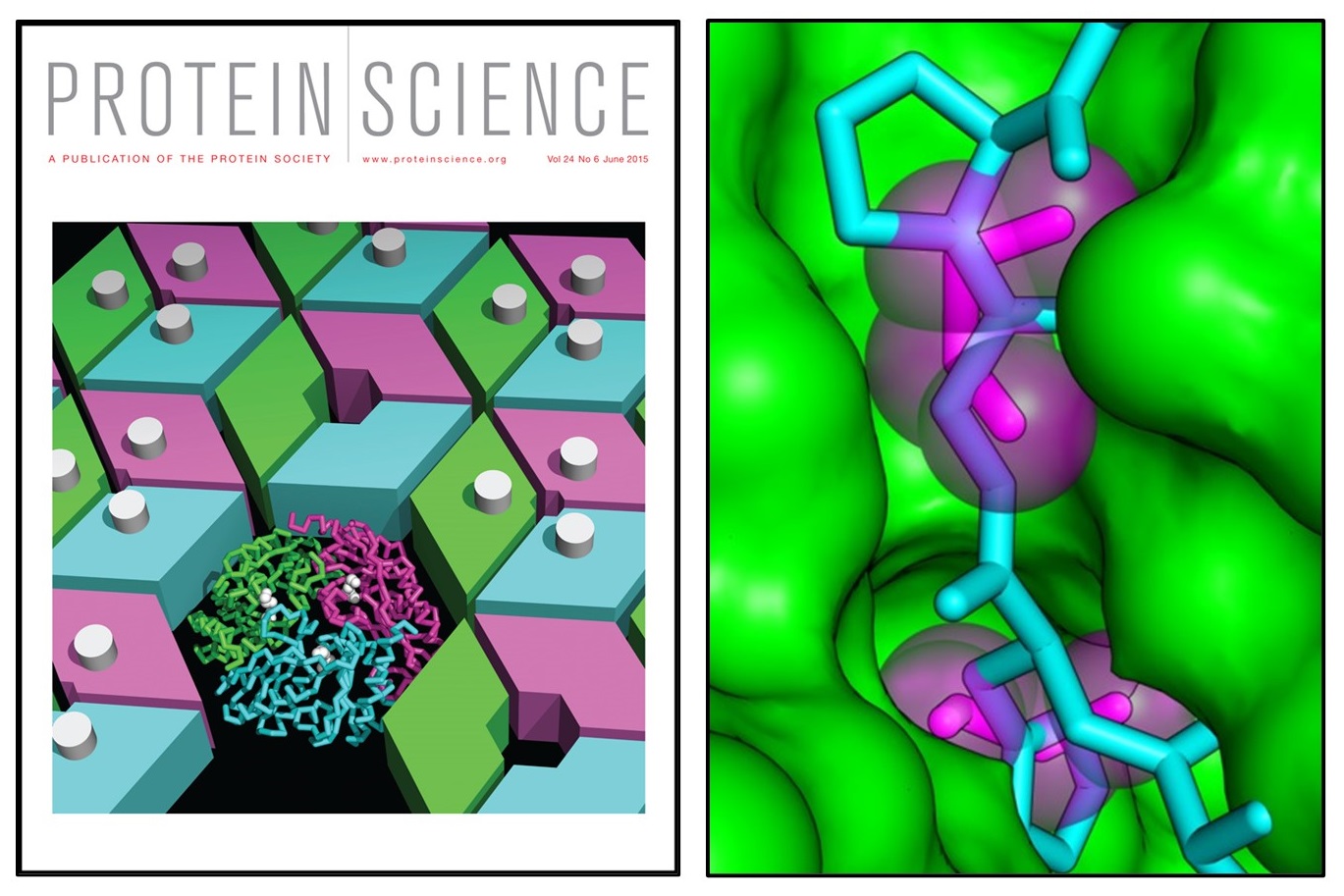
- PMID: 25752492
- PMCID: PMC4456109
- Full Text
- Deposited Structures: 4EDI, 4FDZ, 4TME, 4TM6
- Protein Science Best Paper of 2015 Award
- Yeates lab
Mapping the Conformational Landscape of a Dynamic Enzyme by Multitemperature and XFEL Crystallography
Keedy DA, Kenner LR, Warkentin M, Woldeyes RA, Hopkins JB, Thompson MC, Brewster AS, Van Benschoten AH, Baxter EL, Uervirojnangkoorn M, McPhillips SE, Song J, Alonso-Mori R, Holton JM, Weis WI, Brunger AT, Soltis SM, Lemke H, Gonzalez A, Sauter NK, Cohen AE, van den Bedem H, Thorne RE, Fraser JS. (2015) eLife.
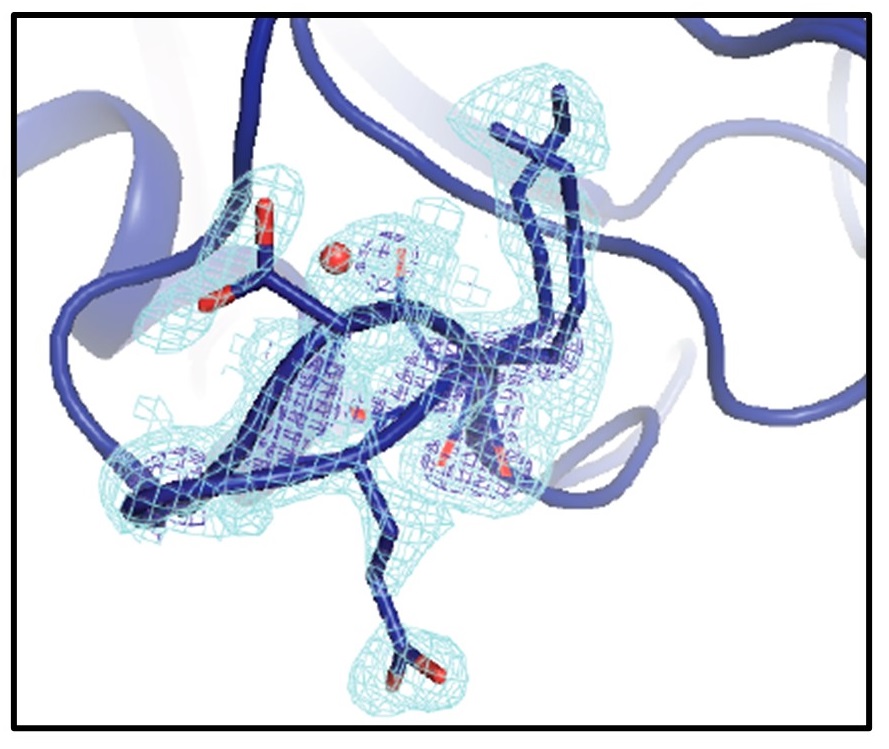
- PMID: 26422513
- PMCID: PMC4721965
- Biorxiv Preprint: 7643
- Full Text
- Online Dataset: doi:10.15785/SBGRID/68
- Deposited Structures: 4YUG, 4YUH, 4YUI, 4YUJ, 4YUK, 4YUL, 4YUM, 4YUN, 4YUO, 4YUP
- Fraser lab @ UC San Francisco
- Paper Submission Celebration Photo
- LCLS XPP Beamline
- Henry van den Bedem
- Thorne lab at Cornell
Structure of a bacterial microcompartment shell protein bound to a cobalamin cofactor
Thompson MC, Crowley CS, Kopstein J, Bobik TA, Yeates TO. (2014) Acta Cryst F.
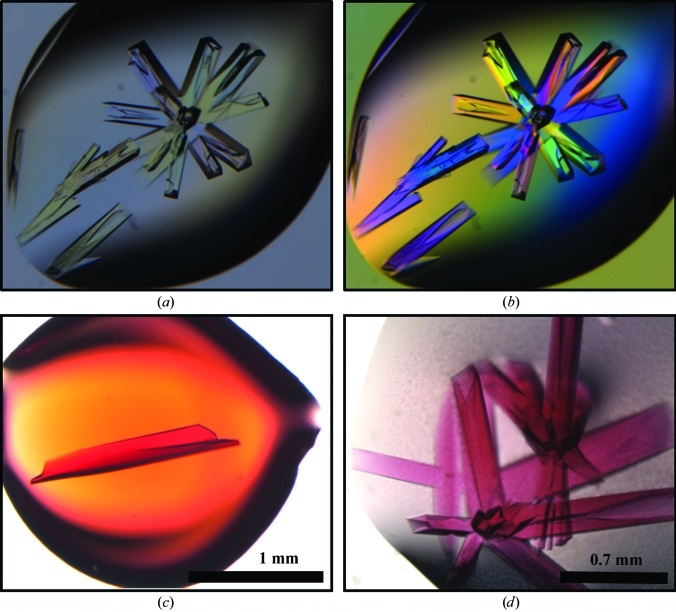
- PMID: 25484204
- PMCID: PMC4259218
- Full Text
- Deposited Structure: 4U6I
- Yeates lab @ UCLA
- Bobik lab @ Iowa State U
Identification of a unique Fe-S cluster binding site in a glycyl-radical type microcompartment shell protein
Thompson MC, Wheatley NM, Jorda J, Sawaya MR, Gidaniyan SD, Ahmed H, Yang Z, McCarty KN, Whitelegge JP, Yeates TO. (2014) J. Mol. Bio.
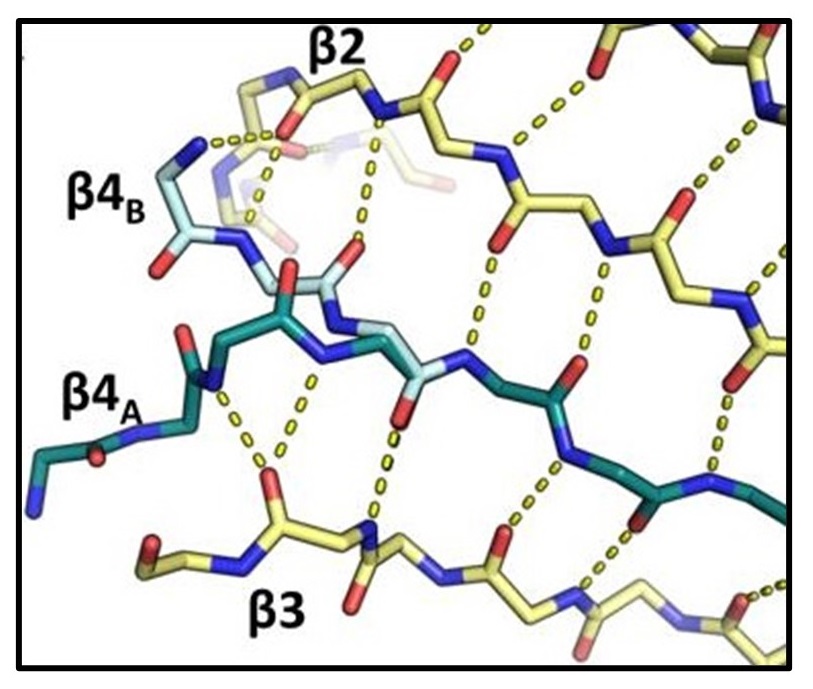
- PMID: 25102080
- PMCID: PMC4175982
- Full Text
- Deposited Structures: 4OLO, 4OLP
- Yeates lab
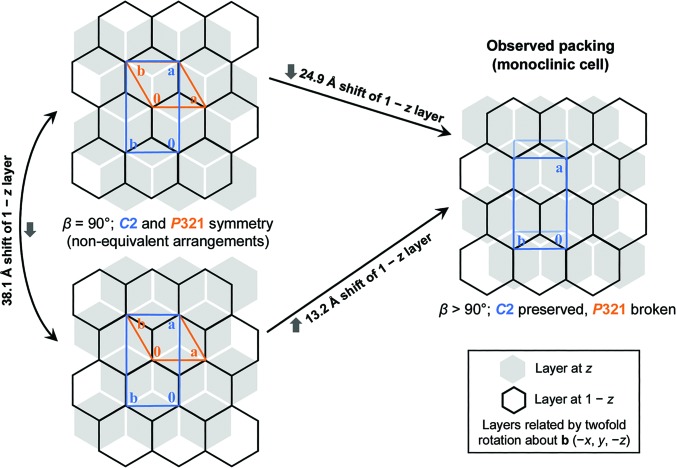
A challenging interpretation of a hexagonally layered protein structure
Thompson MC, Yeates TO. (2013) Acta Cryst D.
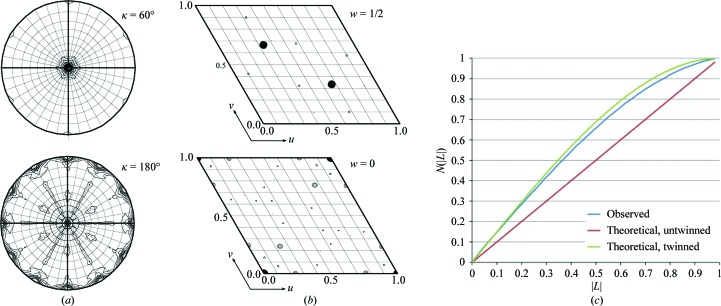
- PMID: 24419393
- PMCID: PMC4030663
- Full Text
- Deposited Structure: 4LIW
- Yeates lab
The protein shells of bacterial microcompartment organelles
Yeates TO, Thompson MC, Bobik TA. (2011) Curr. Opin. Struct. Bio..
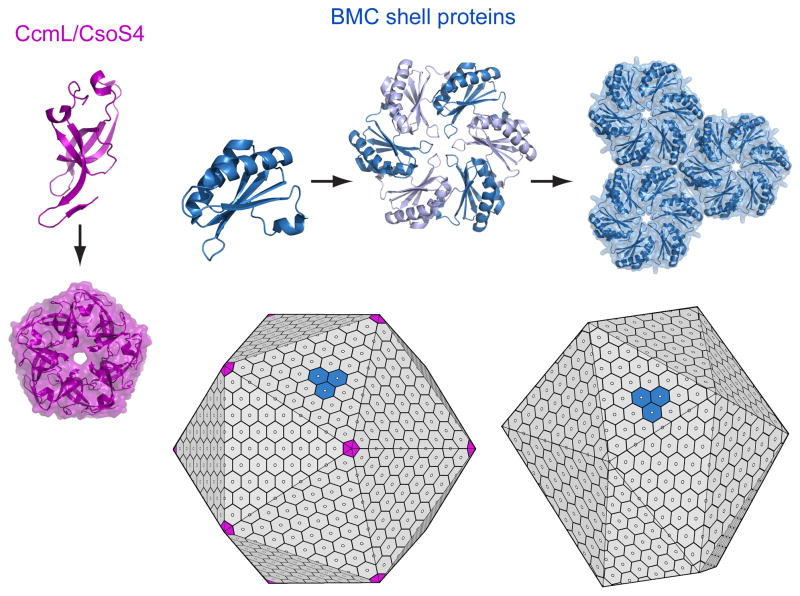
- PMID: 21315581
- PMCID: PMC3070793
- Full Text
- Yeates lab
- Bobik lab @ Iowa State U