Research in the Thompson Lab is focused on understanding the relationship between the structure and function of biological macromolecules, with a particular emphasis on revealing how temperature modulates this relationship.
Temperature is a key variable that governs the behavior of microscopic systems. Contrary to this knowledge, techniques that provide the vast majority of high-resolution structural information about biological macromolecules, including traditional X-ray crystallography and cryoEM, are typically performed using cryogenically preserved samples, in order to minimize the effects of radiation damage. While traditional experiments provide a wealth of structural information about low-energy conformations, cryocooling can bias conformational ensembles and obscure information about fluctuations away from the lowest energy states, which are often critical for function under physiological conditions.
We seek to develop and apply new tools for structural biology that overcome this limitation.
Our research efforts fall into two broad categories:
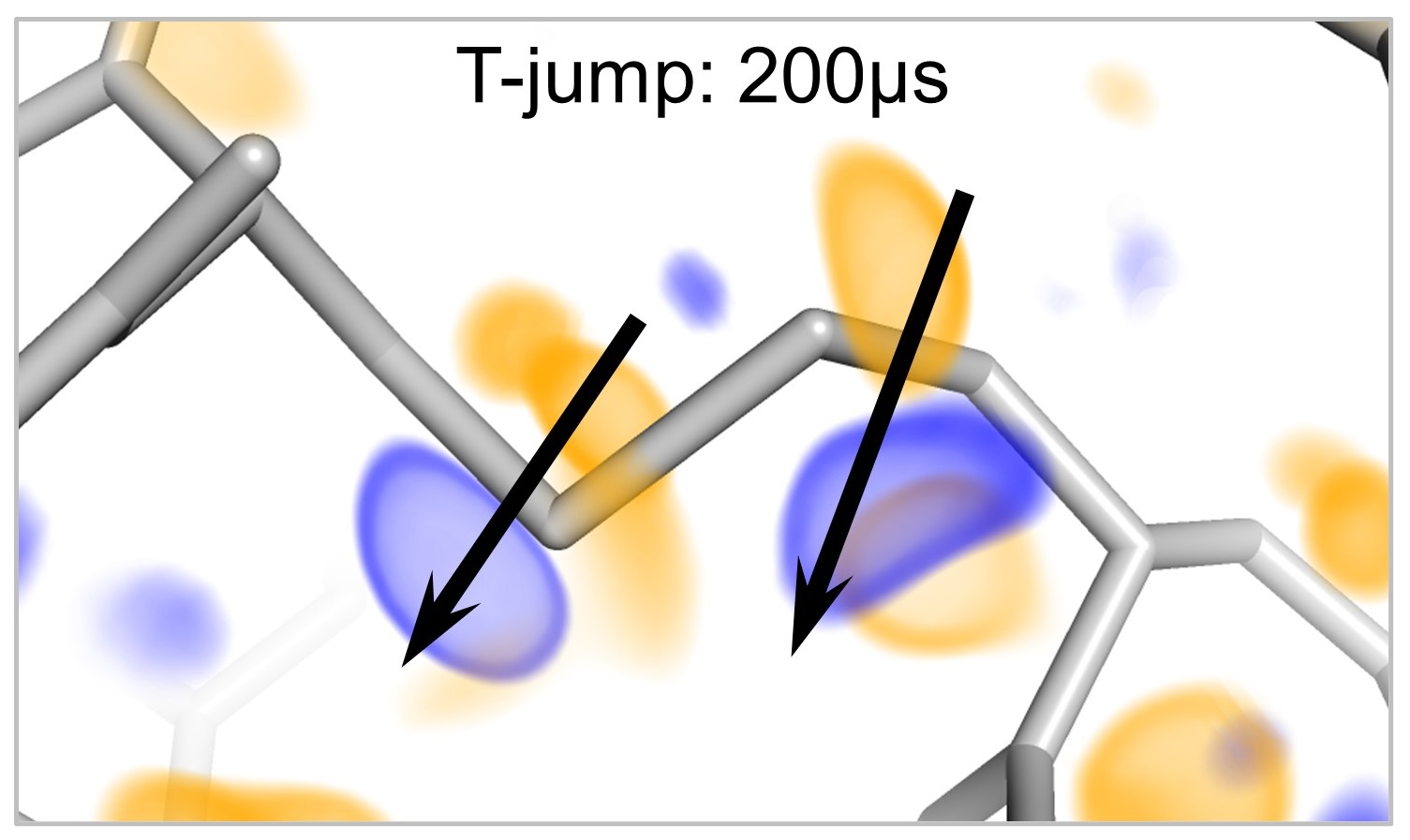
We are developing new experimental methods that utilize temperature perturbations to probe molecular structure and dynamics.
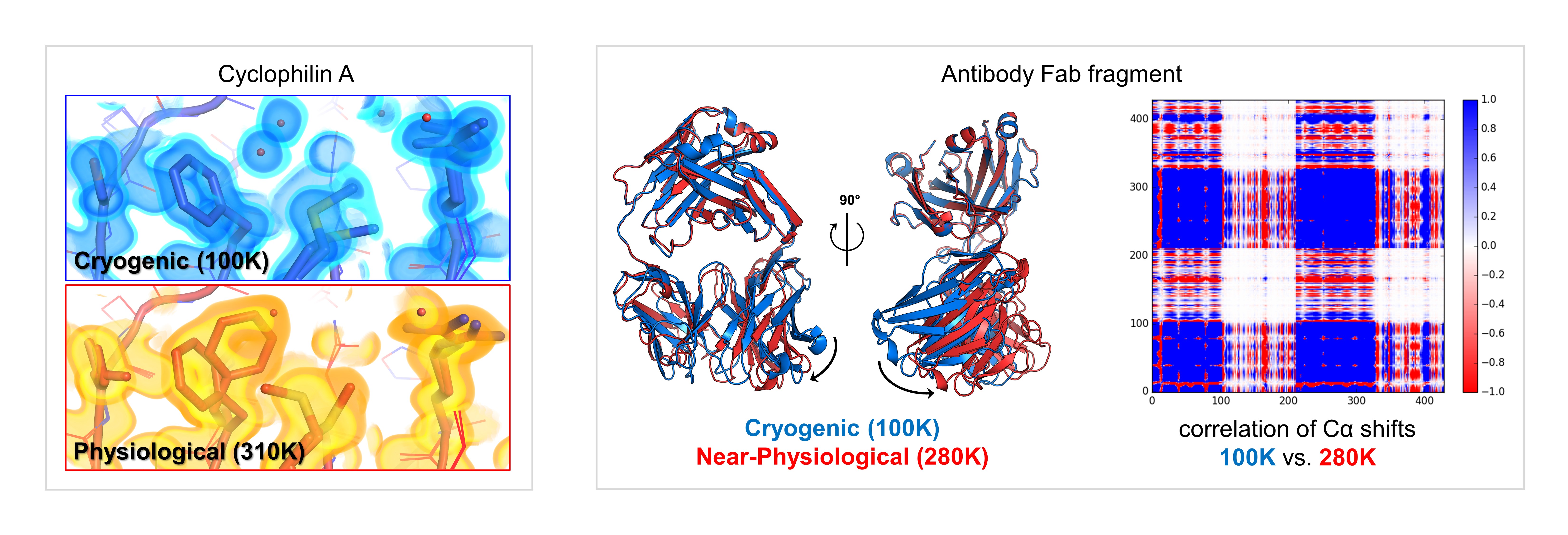
A major goal of our work is to create and democratize instrumentation, data collection protocols, and computational tools to facilitate high-resolution structural biology at non-cryogenic temperatures. We determine the structures of biological macromolecules at their physiologically relevant temperatures, and we use temperature as a general perturbation for static and time-resolved structural measurements. Collectively, these experiments allow us to map the interconversion of conformational states that underlie biochemical phenomena such as enzyme catalysis, ligand binding, and allosteric signal transduction.
Our favorite method is X-ray crystallography, but we are also interested in applying our strategies across a variety of experimental techniques.
We are uncovering the atomic-scale mechanisms of the "molecular thermometers" that allow living systems to sense and respond to temperature.
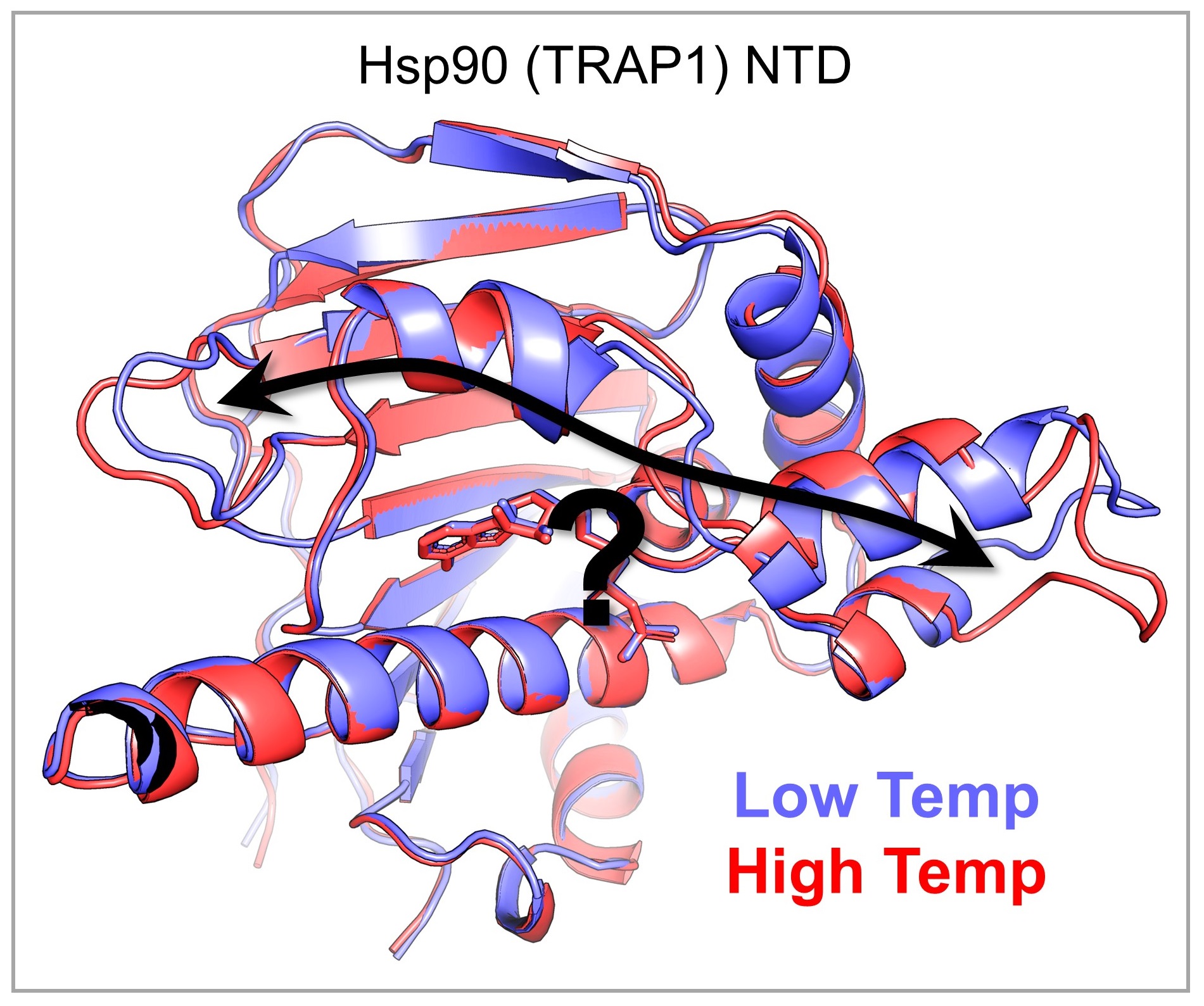
The ability to sense and respond to physiological and environmental temperature variation is pervasive among living systems. Understanding this phenomenon is fundamental to our understanding of life on Earth and how it might be affected by climate change, as well as to our understanding of human health and disease. Genetic and biochemical studies have identified “molecular thermometers,” for which small changes in temperature cause large differences in functional output, but the atomic details of the underlying mechanisms remain poorly understood. To address this knowledge gap, we are determining high-resolution structures of various molecular thermometers across their physiologically relevant temperature ranges. Our observations allow us to generate mechanistic, thermodynamic, and evolutionary hypotheses about the effect of temperature on structure-function relationships, which we test using traditional biochemical and biophysical measurements.
Other projects in the lab focus on advancing protein engineering and drug discovery with structural biology.
We often ask questions at the boundaries of current technical limitations, creating synergy between biological investigations and technology development.
We maintain close collaborations with scientists and engineers at X-ray lightsources around the world.
Our experiments extensively utilize synchrotron beamlines, including BL8.3.1 at the Advanced Light Source (Lawrence Berkeley National Lab), the BioCARS facility at the Advanced Photon Source (Argonne National Lab), and beamlines at the Stanford Synchrotron Radiation Lightsource (SLAC National Accelerator Lab). Additionally, we use X-ray Free Electron Lasers (XFELs) located at the Linac Coherent Light Source (SLAC) and the SPring-8 Angstrom Compact Linear Accelerator (RIKEN, Japan).